Genomics and Epigenomics
Program Overview
The Genomics and Epigenomics Core at our academic institution provides cutting-edge genomic and epigenomic services to researchers. Our facility offers state-of-the-art technologies, expert consultation, and efficient data analysis to advance diverse research projects. We empower scientists to unravel the complexities of genetics and epigenetics, fostering groundbreaking discoveries.
Genomics and Epigenomics Core’s Mission
The mission of the CDI-based branch of the Georgetown Lombardi Comprehensive Cancer Center Genomics & Epigenomics Shared Resource (GESR) is to provide targeted genomic services to researchers. Services in this branch of the GESR are focused on targeted SNP genotyping and DNA methylation analyses, via high-throughput library preparations followed by Nextgen sequencing.
Services and Instruments

 Nextgen targeted sequencing and methyl-seq (bisulfite sequencing): The Fluidigm Juno System uses patented microfluidics technology to perform library preparation for targeted DNA sequencing on the Illumina system using an integrated fluidic circuit (IFC). The dynamic arrays have an integrated network of channels, chambers, and valves that automatically combine the reactions, conserving time reagent and sample amount. Using the Juno system, dozens to hundreds of samples can be processed for targeted Nextgen sequencing of hundreds of amplicons, for SNP and mutation detection, and/or targeted Nextgen Methyl-seq (bisulfite sequencing) for measuring and mapping net and allele-specific CpG methylation. An example of data generated by the Juno MiSEQ pipeline is in Figure 1.
Nextgen targeted sequencing and methyl-seq (bisulfite sequencing): The Fluidigm Juno System uses patented microfluidics technology to perform library preparation for targeted DNA sequencing on the Illumina system using an integrated fluidic circuit (IFC). The dynamic arrays have an integrated network of channels, chambers, and valves that automatically combine the reactions, conserving time reagent and sample amount. Using the Juno system, dozens to hundreds of samples can be processed for targeted Nextgen sequencing of hundreds of amplicons, for SNP and mutation detection, and/or targeted Nextgen Methyl-seq (bisulfite sequencing) for measuring and mapping net and allele-specific CpG methylation. An example of data generated by the Juno MiSEQ pipeline is in Figure 1.
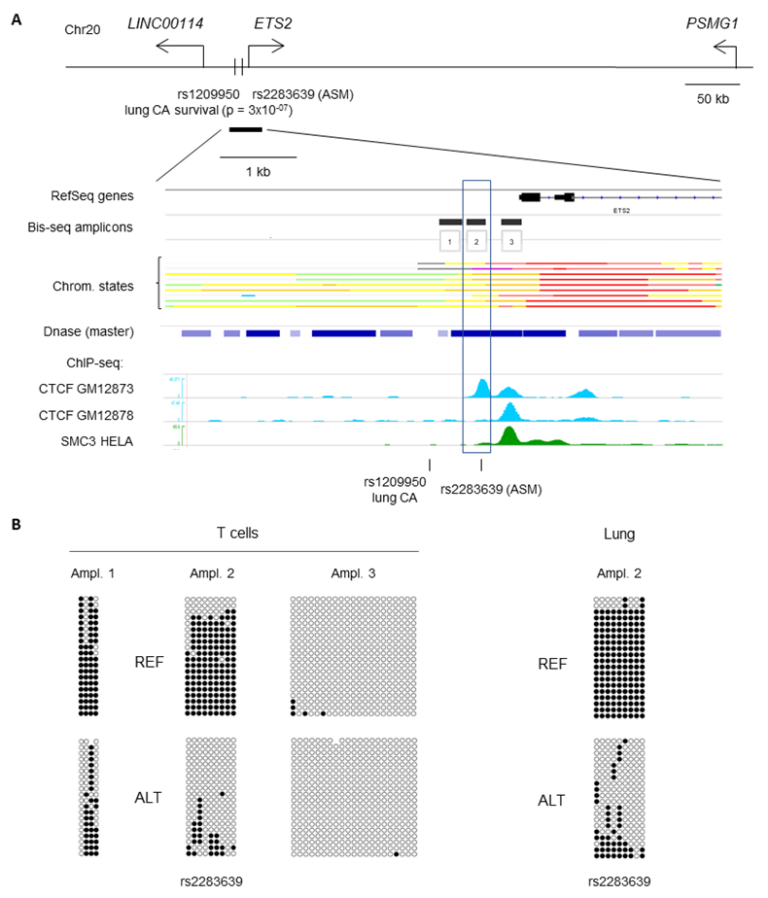
Figure 1. Example of Methyl-seq using the Juno-MiSEQ pipeline. Deep targeted Methyl-seq was utilized to confirm whole-genome bisulfite sequencing data indicating a region of allele-specific CpG methylation (ASM) located within a haplotype block containing a GWAS signal for lung cancer treatment response. A, Map showing the ASM region tagged by index SNP rs2283639, located in a promoter/enhancer region (color-code yellow and red) of the ETS2 gene on chromosome 21. The SNP is close to and in linkage disequilibrium with a GWAS peak SNP rs1209950, associated with survival after treatment of non-small cell lung carcinoma. The ASM index SNP co-localizes with a CTCF ChIP-seq peak in a cluster of multiple TF binding sites and ENCODE ChIP-seq signals. There is also disruption of an ETS1 motif, which could play a role as negative or positive feedback on ETS2 gene expression. Three amplicons were designed for targeted Methyl-seq of the ASM region. B, Results validating a discrete ASM region (amplicon 2) spanning ~600 bp in T cells and lung. Rows represent sequence reads, and columns CpG sites in these reads. All samples are heterozygous, and the reads are separated by allele using SNPs that are not destroyed by the bisulfite conversion. Methylated CpGs are black circles, and unmethylated CpGs are white circles.
Fees and Scheduling
Please complete service request form(s). You will be contacted by the GESR regarding cost and timeline estimates.
Scheduling and billing
Idea Elan core management software will be used for scheduling and billing of services.
Operational Workflow
To ensure efficiency and optimal results, resource members interact closely with investigators. In order to utilize the Shared Resource, users will follow this basic operational workflow:
- An investigator directs an initial inquiry to the GESR Director or Manager or completes the GESR Service Request Form.
- An initial meeting is held during which the feasibility of the project and the overall research plan including data analysis are discussed. The investigator is also notified of the service charges.
- Once deemed feasible, all pertinent information including laboratory personnel dedicated for the project and grant/funding account will be provided by the user through the GESR Sample Submission Form.
- After generating the data, standard reports are produced by the facility, showing quality parameters and net and allele-specific methylation levels and patterns in each biosample.
Investigator completes the Bioinformatics Support Request Form to have the data further analyzed by informatics personnel of the LCCC DS branch of the GESR (optional).
Key Staff/Faculty
Benjamin Tycko, M.D., Ph.D.
Director, Genomics and Epigenomics Core
Dr. Tycko’s lab has a longstanding research program focusing on genetics and epigenetics in human development and disease. Starting with early studies of genomic imprinting and the function of imprinted genes, their work has evolved into genome-wide and locus-specific profiling of CpG methylation, both net and allele-specific, for disease gene and functional variant discovery and pathway analysis in autoimmune diseases, neuropsychiatric disorders, Down syndrome, and cancers. In this work, they have a major focus on cis- and trans-acting effects of local genomic sequences and chromosomal copy number on DNA methylation patterns. They are also studying combination therapies with epigenetic drugs in mouse models of cancer.
Angelica Castano, M.D.
Facility Manager, Genomics and Epigenomics Core
Dr. Castano graduated in 2007 from The Medical School Juan N. Corpas, in Bogota, Colombia. Since coming to the U.S.A., she has over 12 years of academic research experience, focusing on brain tumor molecular biology and genetics and epigenetics in human development and cancer. For the past two years, she has been working on the design, workflow, management, and analysis of Methyl-seq data, using the Juno-MiSEQ pipeline.
